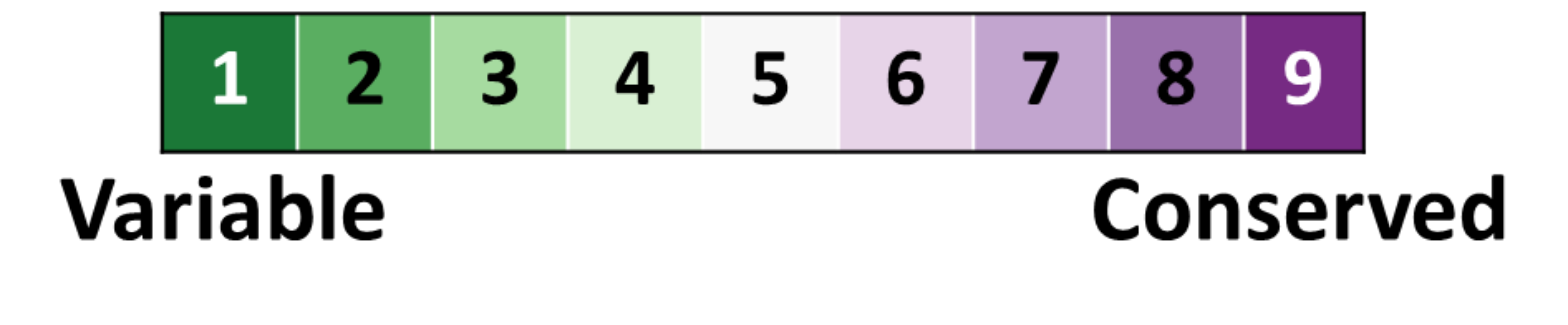
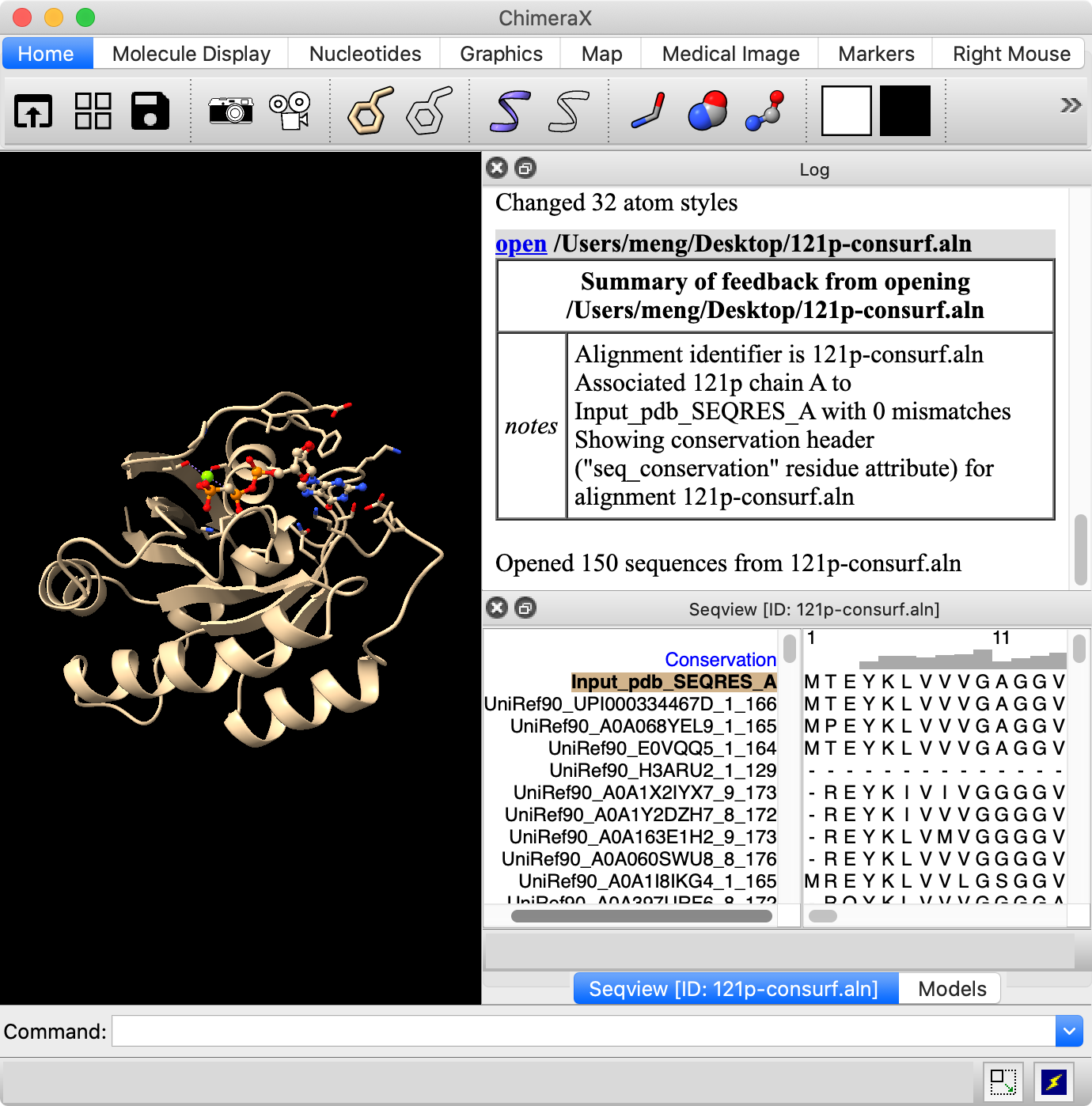
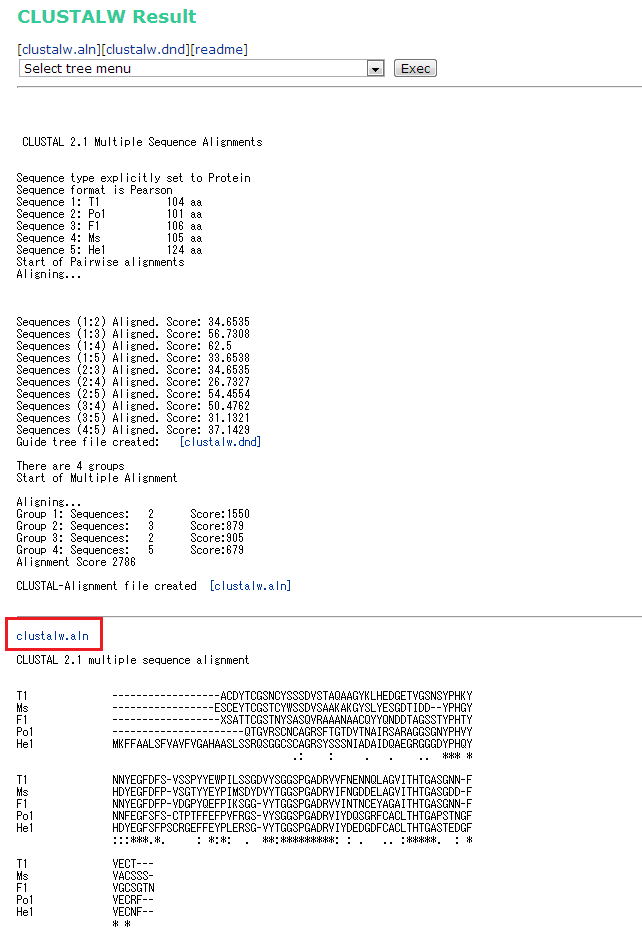
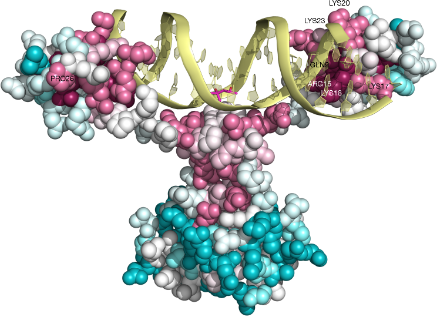
In silico analysis of the tryptophan hydroxylase 2 (TPH2) protein variants related to psychiatric disorders | PLOS ONE
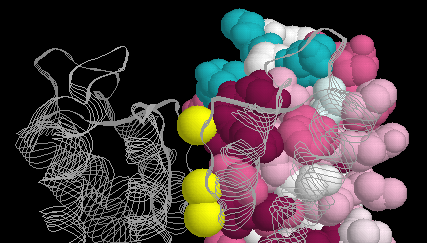
CONSURF analysis of pVirB8 AT (residues 92-231 of structural model).... | Download Scientific Diagram

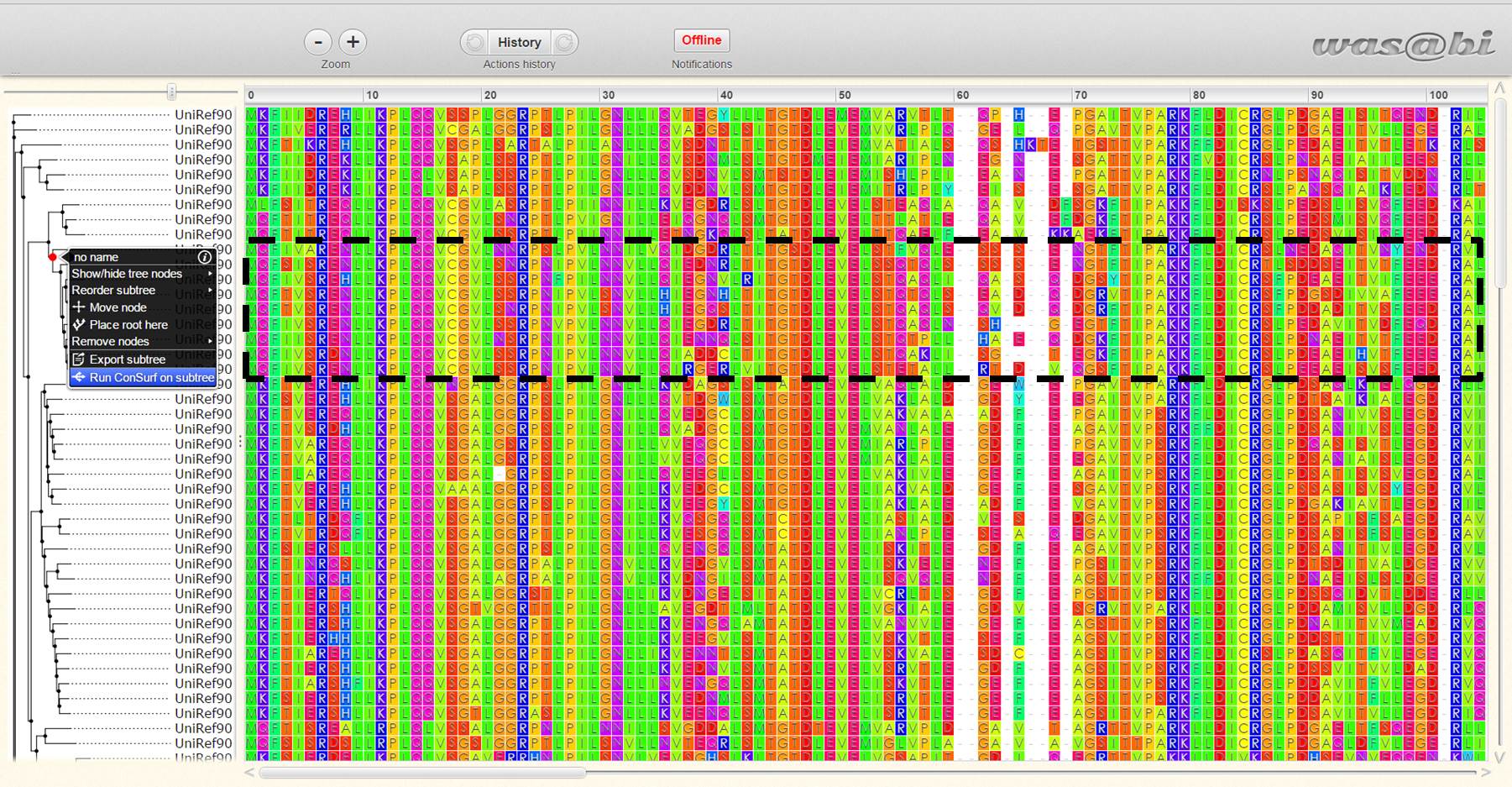
Comparison of conserved amino acid residues. (A) ConSurf server MSA... | Download Scientific Diagram
![PDF] ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function | Semantic Scholar PDF] ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/ebaf71f9bb5f5121064f0cabcb63a8de79b45f20/3-Figure1-1.png)
PDF] ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function | Semantic Scholar
![PDF] ConSurf: Identification of Functional Regions in Proteins by Surface-Mapping of Phylogenetic Information | Semantic Scholar PDF] ConSurf: Identification of Functional Regions in Proteins by Surface-Mapping of Phylogenetic Information | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/f90560898a52a02587229f54950117046f497fdf/2-Figure1-1.png)
PDF] ConSurf: Identification of Functional Regions in Proteins by Surface-Mapping of Phylogenetic Information | Semantic Scholar

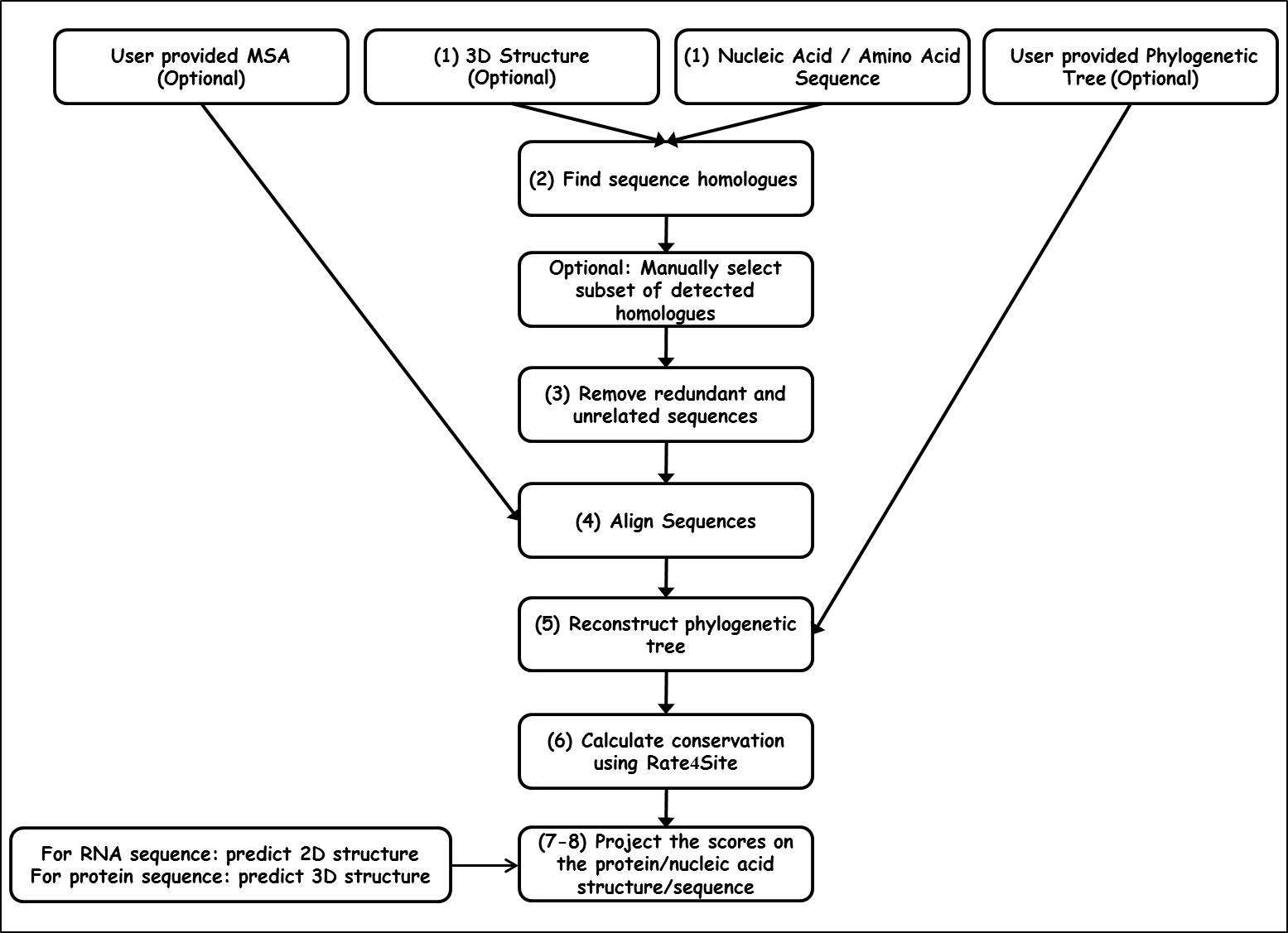
ConSurf‐DB: An accessible repository for the evolutionary conservation patterns of the majority of PDB proteins - Ben Chorin - 2020 - Protein Science - Wiley Online Library

Fig. S2. ConSurf analysis for PpFBPase (A) and PpSBPase (B). Multiple... | Download Scientific Diagram